
Technically not from the BeeCam, but about the same perspective.
After the previous post where I setup a live stream for my beehive, I decided I wanted a quantitative measure of the bee activity going into the winter. The most straightforward thing to start with was simply identifying and counting the bees in each frame of the feed. While not directly measuring anything particularly interesting, this should be well correlated with foraging activity at any time. One might imagine going a step further and identifying attributes about the bees in the image, but I’ll consider that a future improvement for now. A lot of time has passed since the initial development (the year flew by!), but now that I have decent backlog of data, it’s a good time to document my process for counting bees in a video feed.
Having done quite a bit of computer vision in the past, I knew that if I could identify positions in a video frame that were near a bee, I could cluster and count these position estimates to arrive at a total count of bees. The clustering and counting is well suited for the library OpenCV, which provides a very nice image and video API as well. Identifying whether a position is near a bee or not is in total an image recognition or image segmentation task.
U-net inspired neural network to find bees
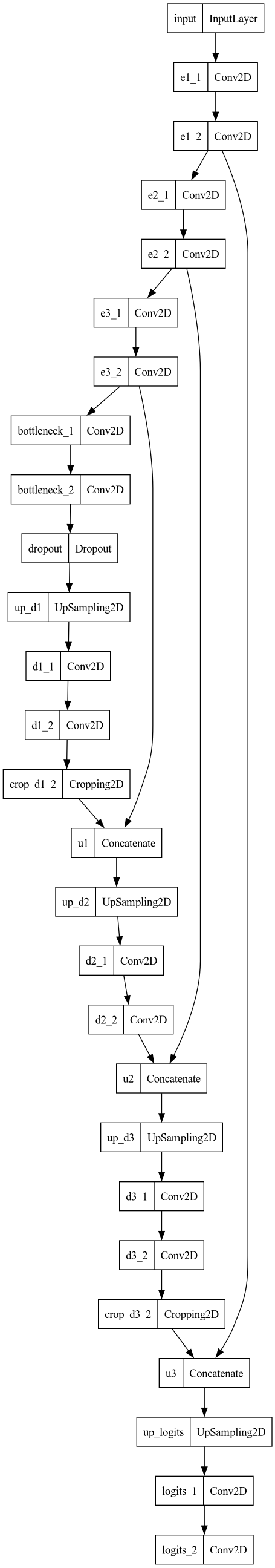
The U-net inspired neural network topology for the BeeAI. Note the skip connections & bottleneck.
In principle methods in OpenCV can tackle this, but modern techniques utilize deep learning and artificial neural networks for image segmentation, to great success. True image recognition as provided by the likes of Resnet-50 would be overkill in this situation. I’m looking more for an idiot savant that can identify bees and only bees than a neural network that can distinguish dog breeds. So, the desired input here would be something like an image frame, and the output would be a probability map of likelihood to be a bee; a sort of image-translation task.
The U-net fits the bill for being able to transform one “image” (input frame) into another “image” (probability map) as long as we play fast and loose with what we use as a channel in a generalized image. U-net has the interesting property of being fully convolutional, which is good here as it means no part of the image will be treated as ‘special’ with all parts using the same convolutional kernels. It is very similar to an auto encoder in that the input can be very similar to the output, and indeed utilizes the concept of a bottleneck, restricting the information that flows though the middle to incentive efficient and general representations. The auto-encoder-like bottleneck is generated by down-scaling with strides in the convolutions, and up-scaling back to the original input dimensions afterwards. U-net also borrows ideas from residual networks by introducing skip-connections between the tiers of up/down scaling. The combination of all these nice features make U-net an excellent at image segmentation, and has given it a prominent place in modern stable diffusion networks like Midjourney where it serves the purpose of transforming noise into the desired output.
For this project, which I’ll call BeeAI, the U-net inspired neural network architecture that worked best in testing is shown on the right. The remainder of this post will go into how to develop the training data and obtain a working BeeAI.
Obtaining input “images”
The input frame could be as simple as an RGB image frozen in time. However, this is a video feed, and in principle there is more information available looking at data over time than simply data at a point in time. Practically, this is the difference between distinguishing live, moving bees from bees that have expired on the front of the hive. The only real visual difference is that the latter don’t move.
To account for this, I will add a fourth channel to the traditional RGB which will be the difference in gray level relative to some moving average of recent frames. Ultimately the neural network will just see this as an additional index in the tensor. What is manifestly just a rapid brightness change can be interpreted as motion, in a sense, as stationary objects would usually show no rapid brightness change. This allows the network to have some sense of motion even if only processing one frame.
Saving raw data
To make this process precise, and collect the necessary raw data from a sequence of OpenCV images, I employ the following python class:
import matplotlib.pyplot as plt
import numpy as np
import cv2, queue, threading, time, collections, itertools
class RawDataProcessor:
def __init__(self):
self.lookback = 100 # Frames to skip between moving average and current image
self.totalhist = 300 # Moving average over the tail end of this, defined by lookback
self.frames = collections.deque(maxlen=self.totalhist)
self.last = None
def process(self,img):
'''Call for every img in a sequence to build the background consistently.'''
self.last = img
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
self.frames.append(gray)
def save(self,prefix='raw/'):
'''At any point in the sequence being `process`ed, generate and save some raw input data.'''
if len(self.frames) < 300:
return # Not ready for background calculation
now_str = datetime.now().strftime('%Y-%m-%d_%H:%M:%S')
cv2.imwrite(f"{prefix}{now_str}_raw.png",self.last)
cv2.imwrite(f"{prefix}{now_str}_gray.png",self.frames[-1])
fdata = np.asarray(list(itertools.islice(self.frames,None,len(self.frames)-self.lookback)))
average_frame = np.mean(fdata, axis=0)
cv2.imwrite(f"{prefix}{now_str}_bg.png",average_frame)
plt.figure(figsize=[14,5])
plt.subplot(1,2,1)
plt.imshow(self.last[...,[2,1,0]])
plt.subplot(1,2,2)
plt.imshow(average_frame)
clear_output(wait=True)
plt.show()
plt.close()
The data collection process is pretty straightforward then: read and process images from a video stream,
from cv2 import VideoCapture
stream = VideoCapture('/path/to/beecam.m3u8')
while saving data periodically,
fp = RawDataProcessor()
skip = 1 # only process every `skip+1`th frame
i = 0 # processed frames
j = 0 # total frames
start_ts = datetime.now()
while True:
try:
for _ in range(skip+1):
res, dat = stream.read()
j = j+1
i = i+1
except Exception as e:
print(e)
time.sleep(10)
stream = VideoCapture(stream_src)
continue
# process every other frame (skip = 1) in a 25 fps stream
fp.process(dat)
# i increases at 12.5 fps, 750 frames is one image a minute (relatively uncorrelated fame-to-frame)
if i%750 == 0:
fp.save()
print('\r',i,j,j/(datetime.now()-start_ts).seconds,end='')
and then sit back and relax!
The RGB image on the left, and the averaged background gray level on the right. Notice how the bees in the shadow of the entrance don’t appear in the background. Took some tweaking of rates and delays to get this to work reliably given how long bees tend to stand still.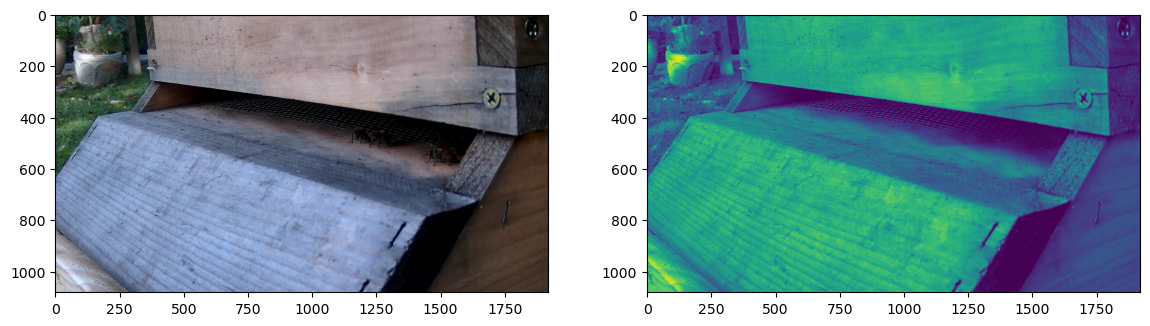
Loading augmented input
As mentioned above, I’ll go a step further than just providing the background to the network, and go ahead with computing the current gray levels and subtracting the two. This will tend to make even poorly defined bees have a measurable signal. I’ll also downsample at this time, and put the data in a sane near-zero order-one range, to avoid having a huge neural network needing many convolutional layers. A raw data loader for input to the network would then be -
INPUT_SIZE = (854,480)
def raw_data_loader(filename):
image = cv2.imread(f"raw/{filename}")
image = cv2.resize(image, INPUT_SIZE)
image = np.asarray(image, dtype=np.float32)[...,[2,1,0]]
image = (image - 127) / 128
bg = cv2.imread(f"raw/{filename.replace('_raw','_bg')}")
bg = cv2.resize(bg, INPUT_SIZE)
bg = np.asarray(bg, dtype=np.float32)[...,[2,1,0]]
bg = (np.mean(bg,axis=2) - 127) / 128
delta = np.abs(np.mean(image,axis=2) - bg)
return np.concatenate([image,delta.reshape(delta.shape+(1,))],axis=2)
So, after loading some data,
augmented_input = raw_data_loader(filename)
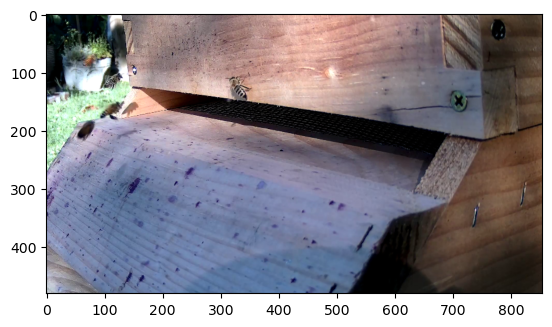
the first three channels would be the RGB part
plt.imshow((augmented_input[...,:-1]+1)/2)
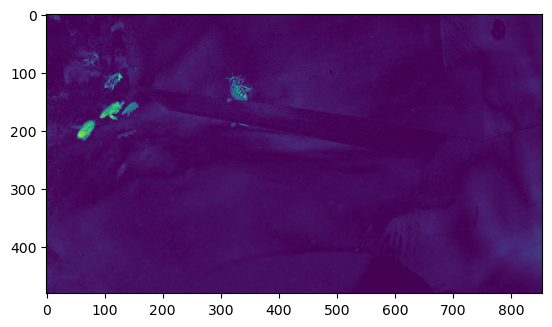
and the last would be the motion.
plt.imshow((augmented_input[...,-1]+2)/4)
Obtaining labeled output “images”
The output is effectively tagged data, and I will have to do the tagging. Generating tagged data with my own effort meant it was time to get organized.
Bee position database
With the following DBee class, I abstract out a SQLite3 database of labeled images, where each image can have multiple labels, consisting of a position on that image (presumably a bee).
import sqlite3
import numpy as np
import pandas as pd
sqlite3.register_adapter(np.int64, lambda val: int(val))
sqlite3.register_adapter(np.int32, lambda val: int(val))
sqlite3.register_adapter(np.uint8, lambda val: int(val))
sqlite3.register_adapter(np.float32, lambda val: float(val))
sqlite3.register_adapter(np.float64, lambda val: float(val))
class DBee:
def __init__(self, filename):
self.con = sqlite3.connect(filename)
cur = self.con.cursor()
cur.execute('''
create table if not exists images (
image_id integer primary key autoincrement,
filename text
)
''')
cur.execute('''
create table if not exists labels (
image_id integer,
x integer,
y integer,
pollen integer
);
''')
def stats(self):
cur = self.con.cursor()
cur.execute('select count(image_id) from images')
num_images = cur.fetchone()[0]
df = pd.read_sql_query("""
select image_id, count(image_id) as num_bees from labels group by image_id
""", self.con)
print('Images:', num_images)
print('Bees: ', df.num_bees.sum())
print(f'Bees/Image: {df.num_bees.sum()/num_images:0.2f}')
def get_filenames(self):
c = self.con.cursor()
return [r[0] for r in c.execute('''select filename from images''').fetchall()]
def get_labels(self, img):
c = self.con.cursor()
c.execute("""
select
l.x,
l.y
from labels l
join images i on l.image_id = i.image_id
where i.filename = ?
""", (img,))
return c.fetchall()
def set_labels(self, img, labels):
img_id = self._id_for_img(img)
if img_id is None:
img_id = self._create_row_for_img(img)
else:
self._delete_labels_for_img_id(img_id)
self._add_rows_for_labels(img_id, labels)
def _id_for_img(self, img):
c = self.con.cursor()
c.execute("select image_id from images where filename=?", (img,))
res = c.fetchone()
if res is None:
return None
else:
return res[0]
def _create_row_for_img(self, img):
c = self.con.cursor()
c.execute("insert into images (filename) values (?)", (img,))
self.con.commit()
return self._id_for_img(img)
def _delete_labels_for_img_id(self, img_id):
c = self.con.cursor()
c.execute("delete from labels where image_id=?", (img_id,))
self.con.commit()
def _add_rows_for_labels(self, img_id, labels):
c = self.con.cursor()
data = [ (img_id, x, y) for x, y in labels ]
c.executemany("insert into labels (image_id, x, y) values (?, ?, ?)", data)
self.con.commit()
def reset(self):
c = self.con.cursor()
c.execute('delete from images')
c.execute('delete from labels')
self.con.commit()
def merge(self, other, fname):
'''Merges two datasets. Files in both keep labels from this dataset'''
assert isinstance(other,DBee), 'Can only merge with DBee and derivatives'
files1 = pd.read_sql_query("""select filename from images""", self.con)
files2 = pd.read_sql_query("""select filename from images""", other.con)
all_files = files1.merge(files2, on='filename', how='outer',indicator=True)
print(len(set(all_files.filename)), len(all_files.filename))
dat1 = pd.read_sql_query("""
select filename, x, y from images i join labels l on i.image_id = l.image_id
""", self.con)
dat2 = pd.read_sql_query("""
select filename, x, y from images i join labels l on i.image_id = l.image_id
""", other.con)
combo_db = DBee(fname)
cur = combo_db.con.cursor()
combo_db.reset()
cur.execute('''drop table if exists all_files''')
all_files.to_sql('all_files',combo_db.con)
cur.execute(f'''
insert into images (
filename
)
select
filename
from all_files
''')
cur.execute('''drop table if exists dat1''')
cur.execute('''drop table if exists dat2''')
dat1.to_sql('dat1',combo_db.con)
dat1.to_sql('dat2',combo_db.con)
cur.execute(f'''
insert into labels (
image_id,
x,
y
)
select
images.image_id,
dat.x,
dat.y
from dat1 as dat
join all_files on all_files.filename = dat.filename
join images on images.filename = dat.filename
where all_files._merge != 'right_only'
''')
cur.execute(f'''
insert into labels (
image_id,
x,
y
)
select
images.image_id,
dat.x,
dat.y
from dat2 as dat
join all_files on all_files.filename = dat.filename
join images on images.filename = dat.filename
where all_files._merge == 'right_only'
''')
cur.execute('drop table all_files')
cur.execute('drop table dat1')
cur.execute('drop table dat2')
combo_db.con.commit()
return combo_db
Bee position labeling tool
Now, with a folder full of raw data and a database to store labels, one only needs to create a tool to show an image with the capability to easily add and remove tag positions. This will allow me to generate databases of positions of bees in random images from a directory by clicking them on a screen, as well as visualize existing databases.
#!/usr/bin/env python3
import tkinter as tk
from PIL import Image, ImageTk, ImageDraw
import os
import sqlite3
import random
import re
import glob
from dbee import DBee
import numpy as np
class LabelUI:
def __init__(self, database, image_dir, width, height, add_images=False):
self.label_db = DBee(database)
self.width = width
self.height = height
self.img_dir = 'raw'
if add_images:
self.files = glob.glob(os.path.join(self.image_dir,'*_raw.png'))
random.shuffle(self.files)
else:
self.files = self.label_db.get_filenames()
# TK UI
root = tk.Tk()
root.title("Bee Labeler")
root.bind("<Right>", self.display_next_image)
print("RIGHT next image")
root.bind("<Left>", self.display_previous_image)
print("LEFT previous image")
root.bind("<Up>", self.toggle_bees)
print("UP toggle labels")
root.bind("n", self.display_next_unlabelled_image)
print("n next image with 0 labels")
root.bind("q", self.quit)
print("q quit")
self.canvas = tk.Canvas(root, cursor="tcross")
self.canvas.config(width=width, height=height)
self.canvas.bind("<Button-1>", self.add_bee_event) # left mouse button
self.canvas.bind("<Button-3>", self.remove_closest_bee_event) # right mouse button
self.canvas.pack()
self.label_map = {}
self.bees_on = True
self.file_idx = 0
self.display_new_image()
root.mainloop()
def quit(self, e):
exit()
def add_bee_event(self, e):
if not self.bees_on:
return
self.add_bee_at(e.x, e.y)
def add_bee_at(self, x, y, radius=4):
rectangle_id = self.canvas.create_rectangle(x-radius, y-radius, x+radius, y+radius, fill="red")
self.label_map[(x, y)] = rectangle_id
def remove_bee(self, rectangle_id):
self.canvas.delete(rectangle_id)
def toggle_bees(self, e):
if self.bees_on:
self.tmp_labels = []
for (x, y), rectangle_id in self.label_map.items():
self.remove_bee(rectangle_id)
self.tmp_labels.append((x, y))
self.label_map = {}
self.bees_on = False
else:
for x, y in self.tmp_labels:
self.add_bee_at(x, y)
self.bees_on = True
def remove_closest_bee_event(self, e):
if not self.bees_on:
return
if len(self.label_map) == 0:
return
closest_point = None
closest_sqr_distance = 0.0
for x, y in self.label_map.keys():
sqr_distance = (e.x - x) ** 2 + (e.y - y) ** 2
if sqr_distance < closest_sqr_distance or closest_point is None:
closest_point = (x, y)
closest_sqr_distance = sqr_distance
self.remove_bee(self.label_map.pop(closest_point))
def display_next_image(self, e=None):
if not self.bees_on:
return
self._flush_changes()
self.file_idx += 1
if self.file_idx == len(self.files):
self.file_idx = len(self.files) - 1
self.display_new_image()
def display_next_unlabelled_image(self, e=None):
self._flush_changes()
while True:
self.file_idx += 1
if self.file_idx == len(self.files):
self.file_idx = len(self.files) - 1
break
if len(self.label_db.get_labels(self.files[self.file_idx])) == 0:
break
self.display_new_image()
def display_previous_image(self, e=None):
if not self.bees_on:
return
self._flush_changes()
self.file_idx -= 1
if self.file_idx < 0:
self.file_idx = 0
self.display_new_image()
def _flush_changes(self):
self.label_db.set_labels(self.files[self.file_idx], self.label_map.keys())
def display_new_image(self):
img_name = self.files[self.file_idx]
title = f'{img_name} {self.file_idx+1} of {len(self.files)}'
img = Image.open(self.img_dir + "/" + img_name)
if img.size != (self.width, self.height):
img = img.resize((self.width, self.height))
canvas = ImageDraw.Draw(img)
canvas.text((0, 0), title, fill="red")
self.tk_img = ImageTk.PhotoImage(img)
self.canvas.create_image(0, 0, image=self.tk_img, anchor=tk.NW)
self.label_map.clear() #The above removes all the rectangles
labels = self.label_db.get_labels(img_name)
np_labels = np.asarray(labels)
for x, y in labels:
dist = np.sqrt(np.sum(np.square(np_labels - np.asarray([x,y])),axis=1))
if np.any(np.logical_and(dist > 0,dist < 10)):
print('Close bees!')
self.add_bee_at(x, y)
import argparse
parser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
parser.add_argument("database", type=str)
parser.add_argument("--image-dir", type=str, default='raw')
parser.add_argument("--add", action='store_true', help='add new images to database')
parser.add_argument("--width", type=int, default=1366, help="label coordinate width")
parser.add_argument("--height", type=int, default=768, help="label coordinate height")
opts = parser.parse_args()
LabelUI(opts.database, opts.image_dir, opts.width, opts.height, add_images=opts.add)
Generating bee proximity output image
A database of bee positions in images is necessary, but not quite what we’re asking the network to generate. Indeed, asking any network to generate a correct-length list of something based on some input is a tricky problem to represent. Instead, I’ll simply encode the labels into an image, and have the network use that as its output.
To do this encoding, I’ll generate a black image and add to it a white pixel at each tagged bee location. While this would work, it’s quite unfair to ask the network to pick one single pixel, as I will not be tagging data that accurately. Instead, I’ll blur this pixel out by a reasonable amount with two Gaussian filters.
LABEL_SIZE = (1366,768)
scale = np.asarray(INPUT_SIZE)/np.asarray(LABEL_SIZE)
labels = pd.read_sql_query('select x, y from labels where image_id = ?', db.con, params=(image_id,))
labels = np.asarray(np.round(np.asarray(labels)*scale),dtype=np.int64)
bitmap = np.zeros(augmented_input.shape[:2],dtype=np.float32)
if len(labels) > 0:
bitmap[labels[:,1],labels[:,0]] = 1
bitmap = cv2.GaussianBlur(bitmap,(9,9),5)
bitmap = cv2.GaussianBlur(bitmap,(9,9),5)
bitmap /= np.max(bitmap)
The amount of blurring was unscientifically chosen such that that the resulting image looked somewhat like a probability map of where I would click as the center of a bee. It’ll be on me to figure out how to decode that representation into the list positions I wanted. One method would be to just sum the image, but this might be heavily biased. Something more like a clustering-centroid approach may work better.
Training BeeAI
Before I get into trying to decode the image-based representation of bee positions, it would be prudent to check that the network can produce them from raw inputs reliably. Practically speaking, there’s an upper limit to the amount of data I’m able or willing to tag myself. I estimate few to tens of seconds per image to accurately tag bees. In that ballpark, O(100 images) is going to take me O(1 hour), so, I manually tagged several hundred images of bees.
With that, I can finally generate a training dataset consisting of inputs and outputs for the network.
This python generator labeled_iter will yield a named tuple containing all the necessary data and metadata:
from collections import namedtuple
db = dbee.DBee('currated_bees.sqlite')
LabeledData = namedtuple('LabeledData',['image_id','filename','input','output'])
def labeled_iter():
images = pd.read_sql_query('select * from images', db.con)
for image_id,filename in zip(images.image_id,images.filename):
augmented_input = raw_data_loader(filename)
scale = np.asarray(INPUT_SIZE)/np.asarray(LABEL_SIZE)
labels = pd.read_sql_query('select x, y from labels where image_id = ?', db.con, params=(image_id,))
labels = np.asarray(np.round(np.asarray(labels)*scale),dtype=np.int64)
bitmap = np.zeros(augmented_input.shape[:2],dtype=np.float32)
if len(labels) > 0:
bitmap[labels[:,1],labels[:,0]] = 1
bitmap = cv2.GaussianBlur(bitmap,(9,9),5)
bitmap = cv2.GaussianBlur(bitmap,(9,9),5)
bitmap /= np.max(bitmap)
yield LabeledData(image_id, filename, augmented_input, bitmap)
Augmenting training data
Several hundred input samples is decent, but not enough variation given the size of the input parameter space for the model to generalize very well. Fortunately this is a classic problem in image-processing neural network applications, and automatic augmentation is baked into the Keras API. The only catch here is that anything transforming the topology of the input (rotations, shifts, scales, etc) must also be applied in an identical way to the output, or the augmentation will effectively be scrambling the data! I’ll apply scales, inversions, and rotations here. Shifts are superfluous for a convolutional-only network. To the input image alone, brightness will be fluctuated. There are many other options.
import tensorflow as tf
augment_both = tf.keras.Sequential([
RandomZoom(0.1,0.1),
RandomFlip("horizontal_and_vertical"),
RandomRotation(0.2),
])
augment_input = tf.keras.Sequential([
RandomBrightness(factor=0.05,value_range=[-2,2])
])
There’s now a bit of a song and dance to get a batch of inputs and apply augmentation in an efficient way. This will take two batches of inputs and outputs, apply the augmentation, and return the augmented batches.
def augment(batch_in, batch_out):
batch = np.concatenate([batch_in, np.reshape(batch_out, batch_out.shape+(1,))], axis=3).copy()
with tf.device('gpu:0'):
batch = augment_both(batch,training=True)
return augment_input(batch[...,:-1],training=True), batch[...,-1]
Generating a first-pass model
Now that the data is sorted, up next is creating the U-net inspired model in Keras.
Fortunately, Keras has exactly the right Lego blocks to plug together.
I’ll parameterize the model in terms of the input width and height along with number of kernels in the first convolution.
Ultimately I chose 16 as base_filter_size.
def construct_model(width, height, base_filter_size):
def conv_block(i, name, filters, kernel_size=3, activation='relu'):
c = Conv2D(filters=filters,
kernel_size=kernel_size,
strides=1,
padding='same',
activation=activation,
name=f"{name}_1")(i)
o = Conv2D(filters=filters,
kernel_size=kernel_size,
strides=2,
padding='same',
activation=activation,
name=f"{name}_2")(c)
return o
def deconv_block(i, name, filters, activation='relu'):
up = UpSampling2D(size=2, name=f'up_{name}')(i)
c = Conv2D(filters=filters,
kernel_size=3,
strides=1,
padding='same',
activation=activation,
name=f"{name}_1")(up)
o = Conv2D(filters=filters,
kernel_size=3,
strides=1,
padding='same',
activation=activation,
name=f"{name}_2")(c)
return o
def fix_shape(d, ref_shape):
if d.shape[1:3] != ref_shape[1:3]:
cropping = ((0,d.shape[1]-ref_shape[1]),(0,d.shape[2]-ref_shape[2]))
name,*_ = d.name.split('/')
d = Cropping2D(cropping=cropping, name=f"crop_{name}")(d)
return d
image = Input(shape=(height, width, 4), name='input')
e1 = conv_block(image, 'e1', filters=base_filter_size)
e2 = conv_block(e1, 'e2', filters=2*base_filter_size)
e3 = conv_block(e2, 'e3', filters=4*base_filter_size)
bottleneck = conv_block(e3, 'bottleneck', filters=8*base_filter_size)
bottleneck = Dropout(0.1)(bottleneck)
d1 = deconv_block(bottleneck, 'd1', filters=4*base_filter_size)
d1 = fix_shape(d1, ref_shape=e3.shape)
d1 = concatenate([d1,e3],axis=3,name='u1')
d2 = deconv_block(d1, 'd2', filters=2*base_filter_size)
d2 = fix_shape(d2, ref_shape=e2.shape)
d2 = concatenate([d2,e2],axis=3,name='u2')
d3 = deconv_block(d2, 'd3', filters=base_filter_size)
d3 = fix_shape(d3, ref_shape=e1.shape)
d3 = concatenate([d3,e1],axis=3,name='u3')
logits = deconv_block(d3, 'logits', filters=1, activation=None)
logits = fix_shape(logits, ref_shape=image.shape)
return Model(inputs=image, outputs=logits)
With the model and data, all that remains is training. Simply loading all the data at once and shoving it into the GPU is not a sustainable practice, and technically the data augmentation can generate an infinite amount of data. As the data loader is an iterator, we’ll abuse that to get infinite data in finite memory.
import itertools
def loop(gen_fn):
while True:
yield from gen_fn()
infinite_iter = loop(labeled_iter)
Now, for a model training loop, abusing the iterators even more.
import gc
model = construct_model(INPUT_SIZE[0], INPUT_SIZE[1], 16)
compile_model(model, 0.0003, pos_weight=10.0)
inputs, outputs = itertools.tee(infinite_iter)
epoch_len = 10000
batch_size = 100
try:
for j in range(10):
print('EPOCH',j)
for i in range(0,epoch_len,batch_size):
inputs = np.asarray(list(dat.input for _,dat in zip(range(batch_size),inputs)))
outputs = np.asarray(list(dat.output for _,dat in zip(range(batch_size),outputs)))
batch_in,batch_out = augment(inputs, outputs)
with tf.device('gpu:0'):
model.fit(batch_in, batch_out, epochs=10, batch_size=25)
gc.collect()
except Exception as e:
print(e)
finally:
model.save_weights('model_v1.h5')
gc.collect()
Bootstrapping
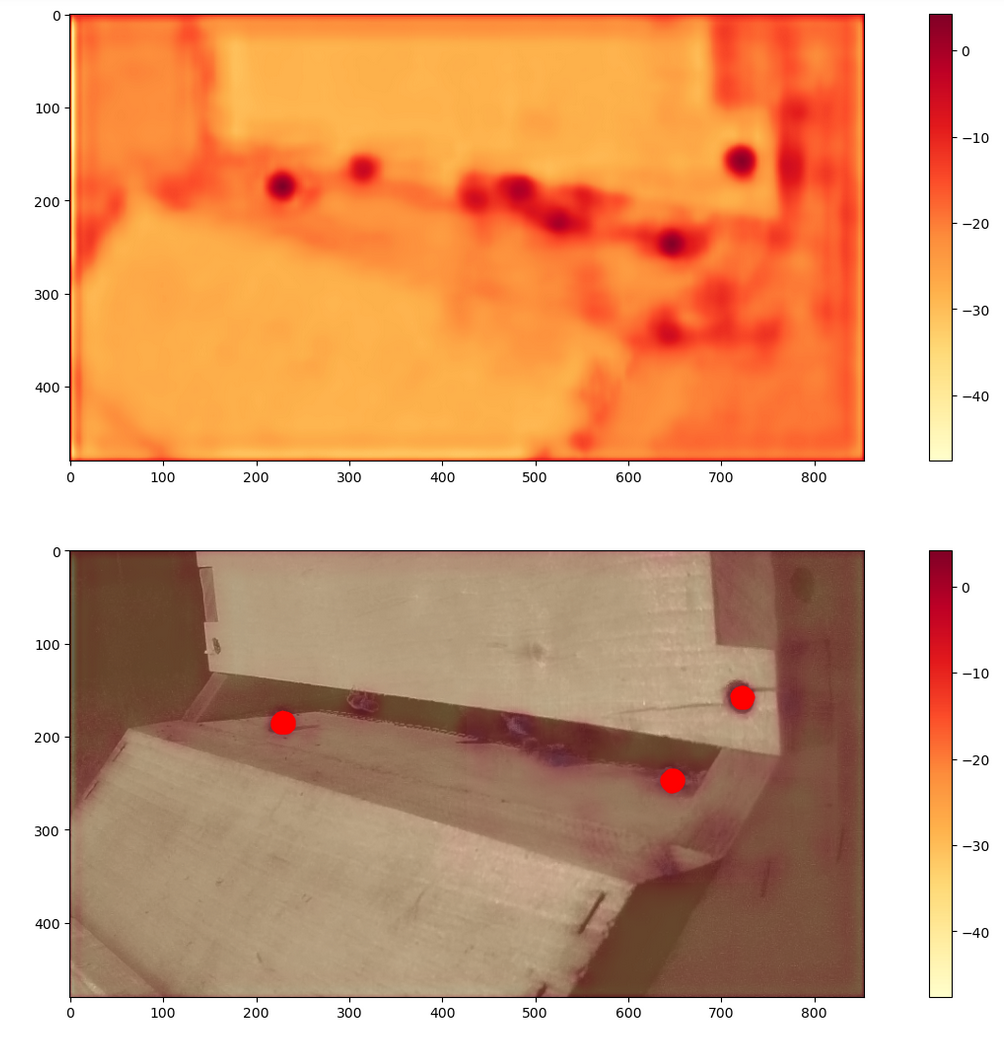
An early look at the v1 model output logits on evaluation data. It does register higher, but not above threshold, where all bees are. It also identifies a screw as a bee. Clearly improvement needed.
Running the training code in the previous section will produce a result. It won’t be great, due to the limited training data. Likely it would be able to tag some bees, but it might overtag or under tag, or perform worse during certain types of weather. The example to the right shows some limitations, where the logits are shown on top, and overlayed with the input image, with above threshold locations marked red.
That said, while it took me tens of seconds to tag an image, this BeeAI can generate output maps for hundreds of images per second. If I only have to review and correct pre-tagged images by BeeAI, I can generate much more labeled training data. Ultimately I repeated the bootstrapping process four times, each time with O(1000) more images until the last, where I deemed the BeeAI was better than myself at tagging bees. For this final step, I allowed the v4 model to label a million images to be used for training v5, and ran the training for 20x as many epochs. I did not verify the labeling of those million images.
After four stages of bootstrapping, the model performs very well, and has very nice tight circles where moving bees appear, with a very smooth background otherwise.
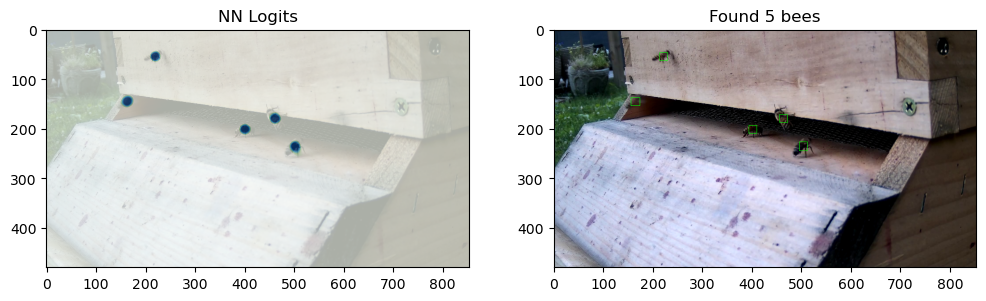
After four stages of bootstrapping, the v5 model is very certain and precise about its bee-centroids.
This, however, begs the question: how did I generate labeled data from the output of BeeAI to do this bootstrapping?
Some computer vision to count clusters
Given the logit image output of the BeeAI neural network,
data = raw_data_loader(fname)
logits = model.predict(np.asarray([data]))[0]
a simple threshold and contour calculation is all it takes to identify a bee and generate the labels for their positions.
thresholded = np.zeros_like(logits,dtype=np.uint8)
thresholded[logits > -0.5] = 255
(contours, _) = cv2.findContours(thresholded.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
labels = []
for cnt in contours:
x,y,w,h = cv2.boundingRect(cnt)
subset = logits[y:y+h,x:x+w]
maxval = np.max(subset)
if maxval > 0.5:
rescale = np.asarray([x+w/2,y+h/2])/INPUT_SIZE*LABEL_SIZE+0.5
labels.append((int(rescale[0]),int(rescale[1])))
Using this, building a bootstrap database is quite straightforward.
Long term data collection
With a trained model and a method for producing bee labels, I need only count the number of produced labels to know how many bees are in any given frame. The remaining difficulty is in putting a Tensorflow model into production. Much to my surprise, there were two distinct major issues with deployment.
First is that out of the box, simply using model.predict appears to leak memory each time its called, to the tune of gigabytes per hour in this case.
The tracemalloc package was clutch for debugging this.
A manual clearing of the global state was required.
Second is that out of the box, Tensorflow appears to assume it can allocate all your GPU memory, and does exactly that. If you’re using your GPU for monitor output (or anything else) this is likely to lock up your entire system if left running, as the graphical session will be unable to resume with no free GPU memory. Strangely, Tensorflow developers appear to be aware of both points, and not too bothered by them.
Something that could run as a system service without taking down the system would look like:
#!/usr/bin/env python3
import numpy as np
from datetime import datetime, timedelta
import cv2, queue, threading, time, collections
import h5py
from IPython.display import Video, clear_output
import sqlite3
import pandas as pd
import keras
from keras.layers import *
from keras.models import Model
from keras.optimizers import Adam
import itertools
import _thread as thread
import time
import gc
import tracemalloc
import sys
import tensorflow as tf
from matplotlib import pyplot as plt
# Fix issue #2
gpus = tf.config.list_physical_devices('GPU')
if gpus:
try:
# Currently, memory growth needs to be the same across GPUs
for gpu in gpus:
tf.config.experimental.set_memory_growth(gpu, True)
logical_gpus = tf.config.list_logical_devices('GPU')
print(len(gpus), "Physical GPUs,", len(logical_gpus), "Logical GPUs")
except RuntimeError as e:
# Memory growth must be set before GPUs have been initialized
print(e)
def construct_model(width, height, base_filter_size):
def conv_block(i, name, filters, kernel_size=3, activation='relu'):
c = Conv2D(filters=filters,
kernel_size=kernel_size,
strides=1,
padding='same',
activation=activation,
name=f"{name}_1")(i)
o = Conv2D(filters=filters,
kernel_size=kernel_size,
strides=2,
padding='same',
activation=activation,
name=f"{name}_2")(c)
return o
def deconv_block(i, name, filters, activation='relu'):
up = UpSampling2D(size=2, name=f'up_{name}')(i)
c = Conv2D(filters=filters,
kernel_size=3,
strides=1,
padding='same',
activation=activation,
name=f"{name}_1")(up)
o = Conv2D(filters=filters,
kernel_size=3,
strides=1,
padding='same',
activation=activation,
name=f"{name}_2")(c)
return o
def fix_shape(d, ref_shape):
if d.shape[1:3] != ref_shape[1:3]:
cropping = ((0,d.shape[1]-ref_shape[1]),(0,d.shape[2]-ref_shape[2]))
name,*_ = d.name.split('/')
d = Cropping2D(cropping=cropping, name=f"crop_{name}")(d)
return d
image = Input(shape=(height, width, 4), name='input')
e1 = conv_block(image, 'e1', filters=base_filter_size)
e2 = conv_block(e1, 'e2', filters=2*base_filter_size)
e3 = conv_block(e2, 'e3', filters=4*base_filter_size)
bottleneck = conv_block(e3, 'bottleneck', filters=8*base_filter_size)
bottleneck = Dropout(0.1)(bottleneck)
d1 = deconv_block(bottleneck, 'd1', filters=4*base_filter_size)
d1 = fix_shape(d1, ref_shape=e3.shape)
d1 = concatenate([d1,e3],axis=3,name='u1')
d2 = deconv_block(d1, 'd2', filters=2*base_filter_size)
d2 = fix_shape(d2, ref_shape=e2.shape)
d2 = concatenate([d2,e2],axis=3,name='u2')
d3 = deconv_block(d2, 'd3', filters=base_filter_size)
d3 = fix_shape(d3, ref_shape=e1.shape)
d3 = concatenate([d3,e1],axis=3,name='u3')
logits = deconv_block(d3, 'logits', filters=1, activation=None)
logits = fix_shape(logits, ref_shape=image.shape)
return Model(inputs=image, outputs=logits)
INPUT_SIZE = (854,480)
LABEL_SIZE = (1366,768)
#tracemalloc.start()
BEGIN = datetime.now()
RUNTIME = np.random.random()*15+15
while True:
model = construct_model(INPUT_SIZE[0], INPUT_SIZE[1], 16)
model.load_weights('model_v5.h5')
con = sqlite3.connect('bee_counter_v5.sqlite')
cur = con.cursor()
cur.execute("CREATE TABLE IF NOT EXISTS bee_counter(timestamp, bee_count)")
con.commit()
stream_src = '/path/to/beecam.m3u8'
stream = cv2.VideoCapture(stream_src)
frames = collections.deque(maxlen=300) # running list of recent history
skip = 1
i = 0
j = 0
start_ts = datetime.now()
try:
#snap_start = tracemalloc.take_snapshot()
while True:
if datetime.now()-timedelta(minutes=RUNTIME) >= BEGIN:
sys.exit(0)
for _ in range(skip+1):
res, image = stream.read()
j = j+1
i = i+1
gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
frames.append(gray)
if len(frames) > 200 and i%10 == 0:
print('processing')
raw = cv2.resize(image, LABEL_SIZE)
image = cv2.resize(image, INPUT_SIZE)
image = np.asarray(image, dtype=np.float32)[...,[2,1,0]]
image = (image - 127) / 128
fdata = list(itertools.islice(frames, None, len(frames)-100))
bg = np.mean(np.asarray(fdata), axis=0)
bg = cv2.resize(bg, INPUT_SIZE)
bg = np.asarray(bg, dtype=np.float32)
bg = (bg - 127) / 128
delta = np.abs(np.mean(image,axis=2) - bg)
augmented_input = np.concatenate([image,delta.reshape(delta.shape+(1,))],axis=2)
logits = model.predict(np.asarray([augmented_input]))[0]
thresholded = np.zeros_like(logits,dtype=np.uint8)
thresholded[logits > -4.0] = 255
(contours, _) = cv2.findContours(thresholded.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
labels = []
sizes = []
maxes = []
for cnt in contours:
x,y,w,h = cv2.boundingRect(cnt)
contour = np.zeros_like(logits,np.uint8)
cv2.drawContours(contour,[cnt],0,255,-1)
mask = contour==255
vals = logits[mask]
maxidx = np.argmax(vals)
maxval = vals[maxidx]
y_s,x_s = [dim[maxidx] for dim in np.where(mask)[:2]]
print(maxval)
if maxval > -2.0:
lx,ly = np.asarray([x_s,y_s])/INPUT_SIZE*LABEL_SIZE+0.5
lx,ly = int(lx),int(ly)
if lx != 0 and ly != 0:
labels.append((lx,ly))
sizes.append((w,h))
maxes.append(maxval)
cur = con.cursor()
cur.execute("INSERT INTO bee_counter VALUES (?,?)",(datetime.now(),len(labels)))
if i % 500 == 0:
con.commit()
print(f'fps: {j/(datetime.now() - start_ts).seconds:0.2f}')
gc.collect()
keras.backend.clear_session() # Fix issue #1
#snap_now = tracemalloc.take_snapshot()
#for detail in snap_now.compare_to(snap_start, 'lineno')[:10]:
# print(detail)
print(datetime.now(),len(labels),labels)
except Exception as e:
print(e)
time.sleep(10)
This service creates a nice database table containing the results of running the model continuously over the video feed.
Some results from the recent months
I’ve been collecting data using BeeAI since July 2023, following the hive in its down-sizing into the winter. Bees/Frame (that is, video frame, not hive frame) is not the most intuitive number, but it is exactly what I am measuring. It is roughly proportional to the number of bees entering+exiting the hive at any given time.
Activity has reduced over this period, roughly as one would expect. One can also see the length of the day shorten, corresponding to reduced hive activity periods. Note also that nighttime activity goes away completely as temperatures get into the 50s at night. The bees are still doing well, with good prospects for spring.
Here’s an average day’s activity at weekly intervals.
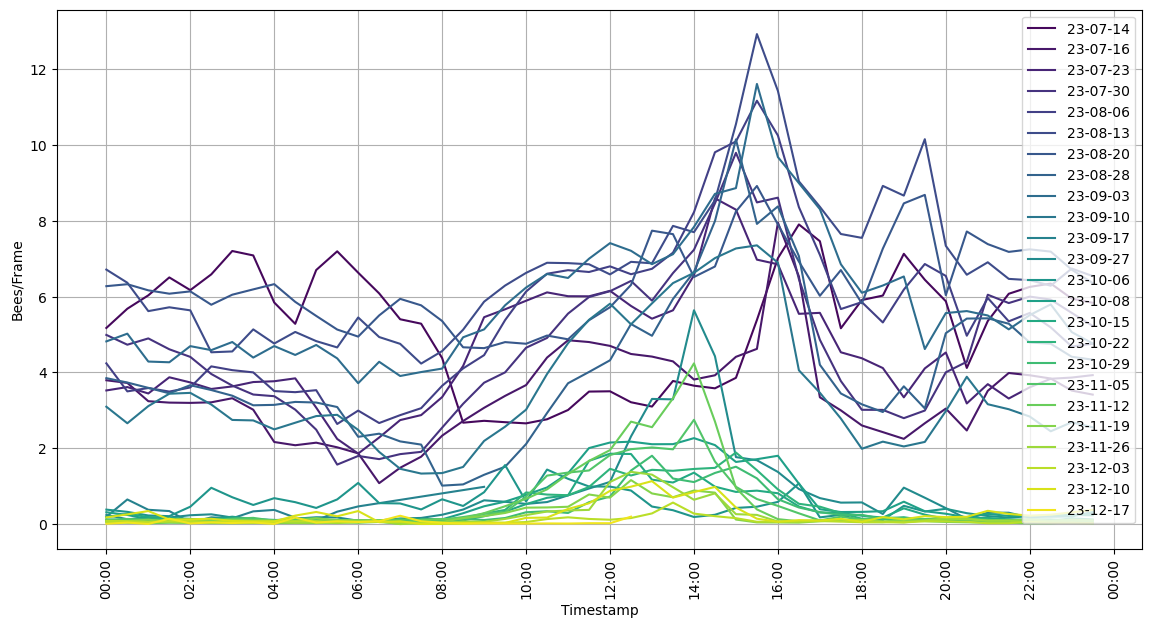
And here is a per-day for each week.
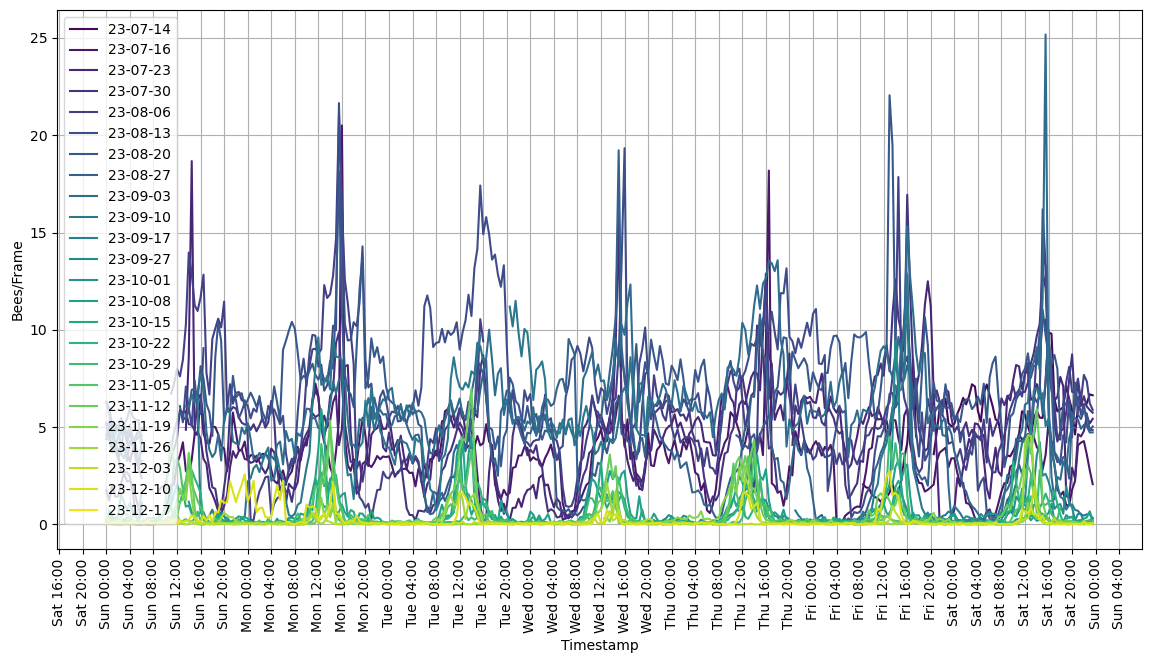
Haven’t yet decided what to do with this data, however it’s been a helpful tool for keeping tabs on the hive activity when I don’t have time to go watch them as frequently as I would like.
>> Home